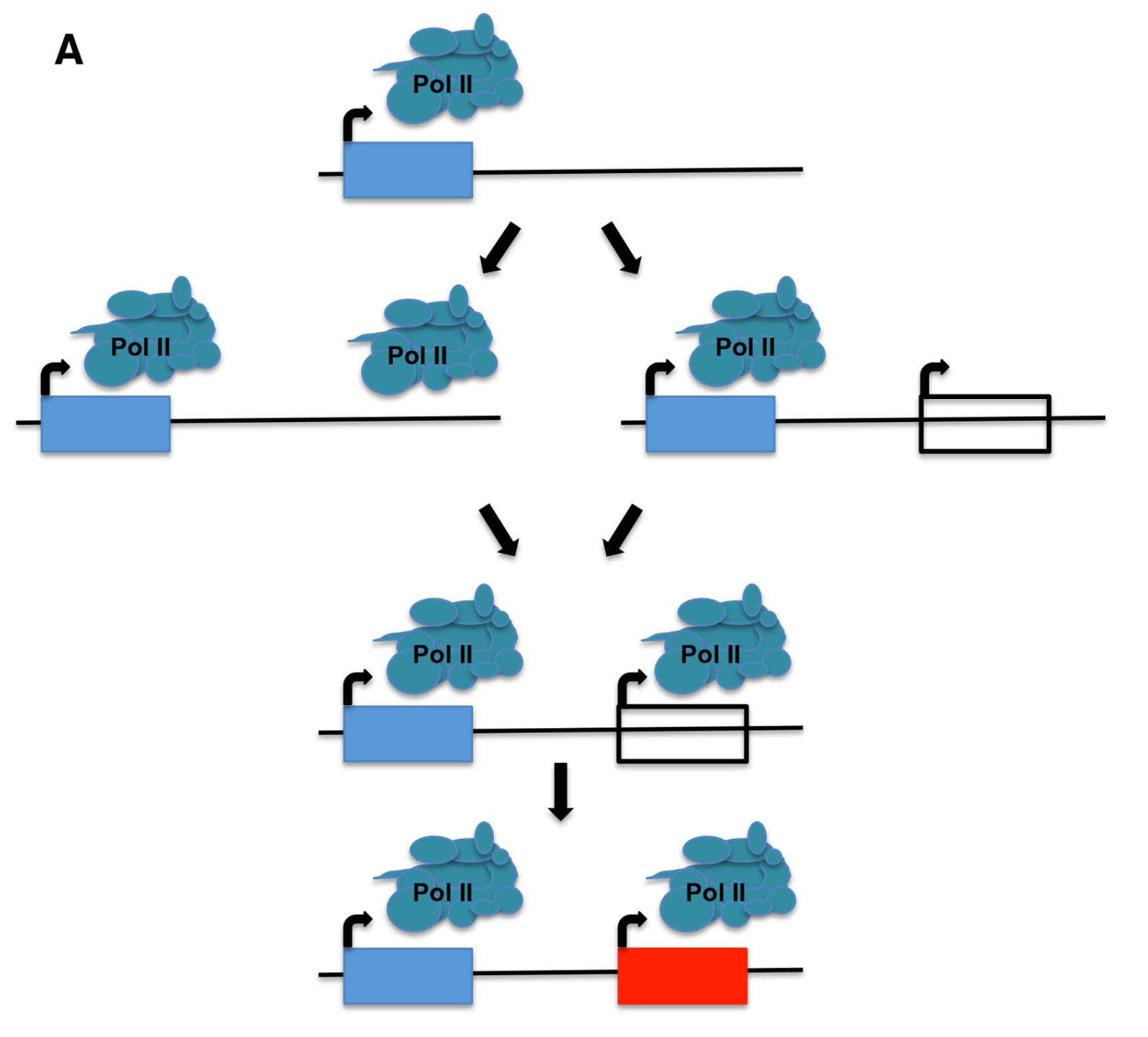
 Community benchmarking and evaluation of human unannotated microprotein detection by mass spectrometry based proteomics
Community benchmarking and evaluation of human unannotated microprotein detection by mass spectrometry based proteomics
Nature Communications (2026)
 Emergence and tandem repeat-mediated elongation of a translated de novo open reading frame in human oncogenic RNA gene VPS9D1-AS1 (MYU)
Emergence and tandem repeat-mediated elongation of a translated de novo open reading frame in human oncogenic RNA gene VPS9D1-AS1 (MYU)
Lin Chou, Shu-Ting Cho, Jiwon Lee, David Laub, Douglas Meyer, Hannah Carter, Anne-Ruxandra Carvunis
Genome Biology and Evolution (2026)
 Translon: a single term for translated regions
Translon: a single term for translated regions
Michał I. Świrski, Jack A. S. Tierney, M. Mar Albà, Dmitry E. Andreev, Julie L. Aspden, John F. Atkins, Michal Bassani-Sternberg, Marla J. Berry, Stefano Biffo, Kathleen Boris-Lawrie, Mark Borodovsky, Ian Brierley, Matthew Brook, Marie A. Brunet, Janusz M. Bujnicki, Neva Caliskan, Lorenzo Calviello, Anne-Ruxandra Carvunis, Jamie H. D. Cate, Can Cenik, Kung Yao Chang, Yiwen Chen, Sonia Chothani, Jyoti S. Choudhary, Patricia L. Clark, Jim Clauwaert, Lynn Cooley, Erik Dassi, Kellie Dean, Jean-Jacques Diaz, Christoph Dieterich, Rivka Dikstein, Jonathan D. Dinman, Sergey E. Dmitriev, Olga A. Dontsova, Christine M. Dunham, Sandeep M. Eswarappa, Philip J. Farabaugh, Pouya Faridi, Ivo Fierro-Monti, Andrew E. Firth, David Gatfield, Fátima Gebauer, Mikhail S. Gelfand, Nicola K. Gray, Rachel Green, Chris H. Hill, Ya-Ming Hou, Norbert Hübner, Zoya Ignatova, Pavel Ivanov, Shintaro Iwasaki, Rory Johnson, Ahmad Jomaa, Marko Jovanovic, Irwin Jungreis, Manolis Kellis, Jeffrey S. Kieft, Alex V. Kochetov, Eugene V. Koonin, Andrei A. Korostelev, Joanna Kufel, Ivan V. Kulakovskiy, Leo Kurian, Denis L. J. Lafontaine, Ola Larsson, Gary Loughran, Julius Lukeš, Marco Mariotti, Elena S. Martens-Uzunova, Thomas F. Martinez, Akinobu Matsumoto, Joel McManus, Jan Medenbach, Sergey V. Melnikov, Gerben Menschaert, Catharina Merchante, Martin Mikl, W. Allen Miller, Oliver Mühlemann, Olivier Namy, Danny D. Nedialkova, Jozef Nosek, Sandra Orchard, Petar Ozretić, Mihaela Pertea, Dmitri D. Pervouchine, Luísa Romão, David Ron, Xavier Roucou, Maria P. Rubtsova, Jorge Ruiz-Orera, Alan Saghatelian, Steven L. Salzberg, Lucia A. Seale, Cathal Seoighe, Petr V. Sergiev, Premal Shah, Nikolay Shirokikh, Sarah A. Slavoff, Nahum Sonenberg, Timothy J. Stasevich, Roman J. Szczesny, Tiina Tamm, Marek Tchórzewski, Ivan Topisirovic, Michel L. Tremblay, Tamir Tuller, Igor Ulitsky, Leoš Shivaya Valášek, Petra Van Damme, Gabriella Viero, Juan Antonio Vizcaino, Christine Vogel, Edward W. J. Wallace, Jonathan S. Weissman, Eric Westhof, Nicola Whiffin, Daniel N. Wilson, Zhi Xie, Jonathan W. Yewdell, Martina M. Yordanova, Chien-Hung Yu, Vyacheslav Yurchenko, Bojan Zagrovic, TRANSLACORE, Eivind Valen & Pavel V. Baranov
Nature Methods (2025)
 An expanded reference catalog of translated open reading frames for biomedical research
An expanded reference catalog of translated open reading frames for biomedical research
Sonia Chothani, Jorge Ruiz-Orera, Jack A. S. Tierney, Jim Clauwaert, Eric W Deutsch, M. Mar Alba, Julie L. Aspden, Pavel V. Baranov, Ariel Alejandro Bazzini, Elspeth A. Bruford, Marie A. Brunet, Tristan Cardon, Anne-Ruxandra Carvunis, Claudio Casola, Jyoti Sharma Choudhary, Kellie Dean, Pouya Faridi, Ivo Fierro-Monti, Isabelle Fournier, Adam Frankish, Mark Gerstein, Norbert Hubner, Yunzhe Jiang, Manolis Kellis, Leron W. Kok, Thomas F Martinez, Gerben Menschaert, Pengyu Ni, Sandra Orchard, Xavier Roucou, Joel Rozowsky, Michel Salzet, Mauro Siragusa, Sarah Slavoff, Michal I Swirski, Nicola Ternette, Eivind Valen, Juan Antonio Vizcaino, Aaron Wacholder, Wei Wu, Zhi Xie, Yucheng T. Yang, Robert L. Moritz, Jonathan Mudge, Sebastiaan van Heesch, John R Prensner, Owen JL Rackham
bioRxiv (2025)
 What is the current bottleneck in mapping molecular interaction networks?
What is the current bottleneck in mapping molecular interaction networks?
Michael A. Skinnider , Katja Luck, M. Shahid Mukhtar, Martin Garrido-Rodriguez, Julio Saez-Rodriguez, Jolanda van Leeuwen, Pedro Beltrao, Anne-Ruxandra Carvunis, Mikko Taipale, Andrew Emili, Martha L. Bulyk, Nevan J. Krogan
Cell Systems (2025)
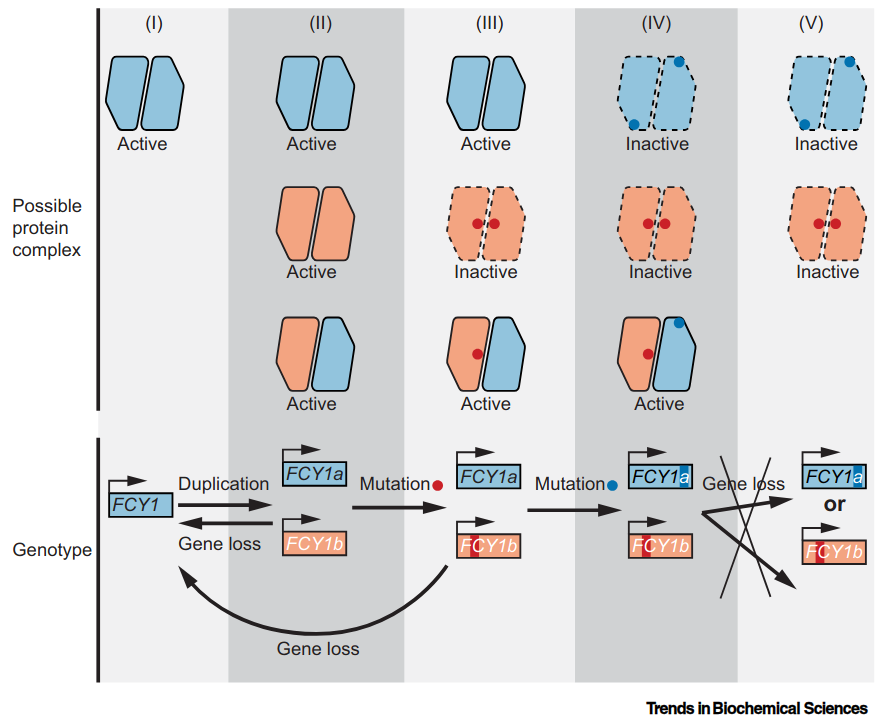 Constructive neutral evolution of homodimer to heterodimer transition
Constructive neutral evolution of homodimer to heterodimer transition
Chou L, Houghton CJ, Wacholder A,Carvunis AR.
Trends in Biochemical Sciences (2024)
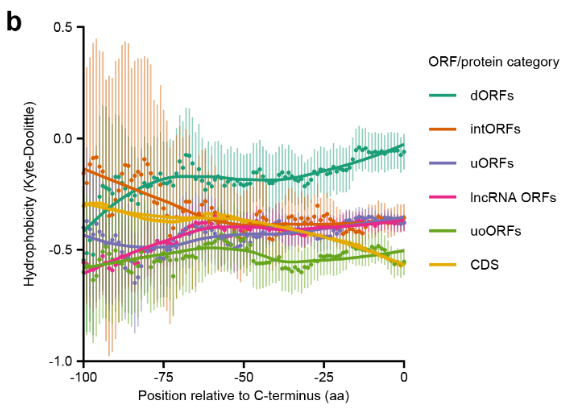 High-quality peptide evidence for annotating non-canonical open reading frames as human proteins
High-quality peptide evidence for annotating non-canonical open reading frames as human proteins
bioRxiv (2024)
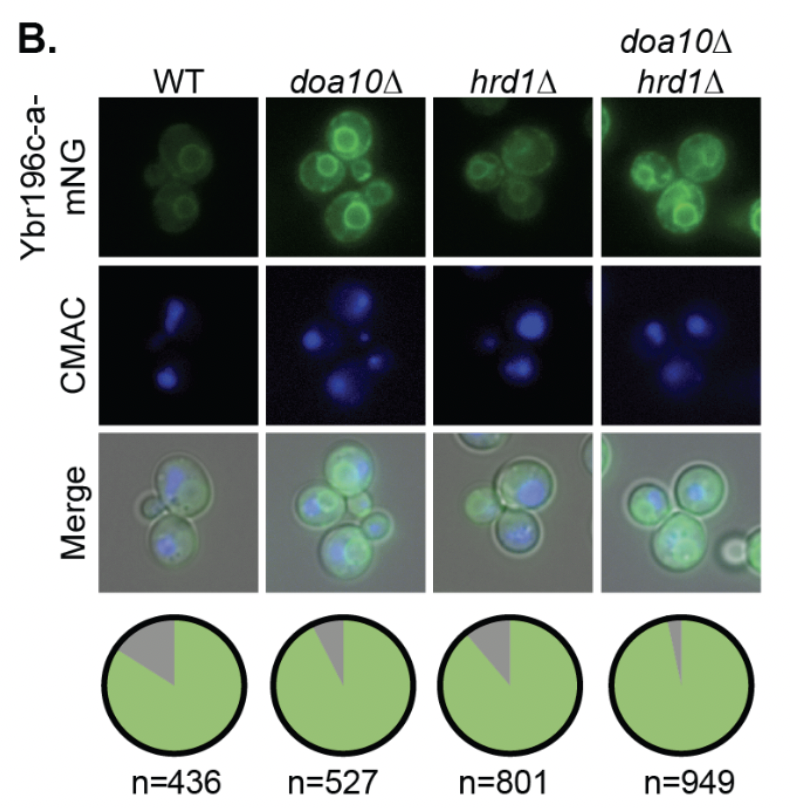 Cellular processing of beneficial de novo emerging proteins
Cellular processing of beneficial de novo emerging proteins
Houghton CJ, Castilho Coelho N, Chiang A, Hedayati S, Parikh SB, Ozbaki-Yagan N, Wacholder A, Iannotta J, Berger A, Carvunis AR, O’Donnell AF.
bioRxiv (2024)
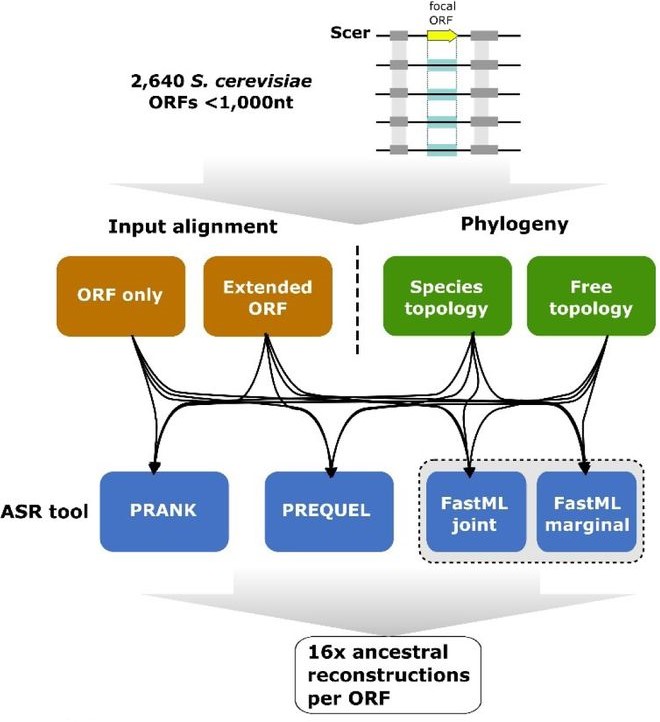 Ancestral Sequence Reconstruction as a Tool to Detect and Study De Novo Gene Emergence
Ancestral Sequence Reconstruction as a Tool to Detect and Study De Novo Gene Emergence
Vakirlis N, Acar O, Cherupally V, Carvunis AR.
Genome Biology and Evolution (2024)
 Massively integrated coexpression analysis reveals transcriptional regulation, evolution and cellular implications of the yeast noncanonical translatome
Massively integrated coexpression analysis reveals transcriptional regulation, evolution and cellular implications of the yeast noncanonical translatome
Rich A, Acar O, Carvunis AR.
Genome Biology (2024)
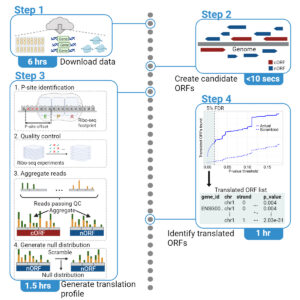 Integrative detection of genome-wide translation using iRibo
Integrative detection of genome-wide translation using iRibo
Turcan A, Lee J, Wacholder A, Carvunis AR.
STAR Protocols (2024)
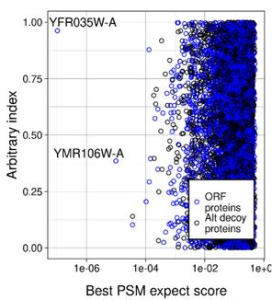 Biological Factors and Statistical Limitations Prevent Detection of Most Noncanonical Proteins by Mass Spectrometry
Biological Factors and Statistical Limitations Prevent Detection of Most Noncanonical Proteins by Mass Spectrometry
Wacholder A & Carvunis AR.
PLOS Biology (2023)
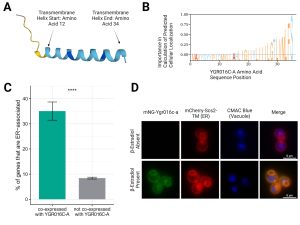 Unannotated Open Reading Frame in Saccharomyces cerevisiae Encodes Protein Localizing to the Endoplasmic Reticulum
Unannotated Open Reading Frame in Saccharomyces cerevisiae Encodes Protein Localizing to the Endoplasmic Reticulum
Chang S*, Joyson M*, Kelly A*, Tang L*, Iannotta J, Rich A, Castilho Coelho N, Carvunis AR.
microPublication Biology (2023)
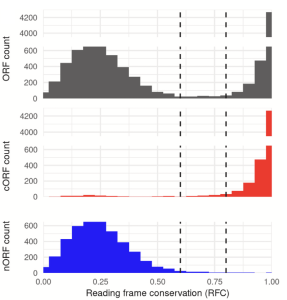 A vast evolutionarily transient translatome contributes to phenotype and fitness
A vast evolutionarily transient translatome contributes to phenotype and fitness
Wacholder A, Parikh SB, Coelho NC, Acar O, Houghton C, Chou L, Carvunis AR
Cell Systems (2023). Link to the perspective written by Zachary Ardern and Md Hassan uz-Zaman in Cell Systems. This work is also recommended in H1 connect.
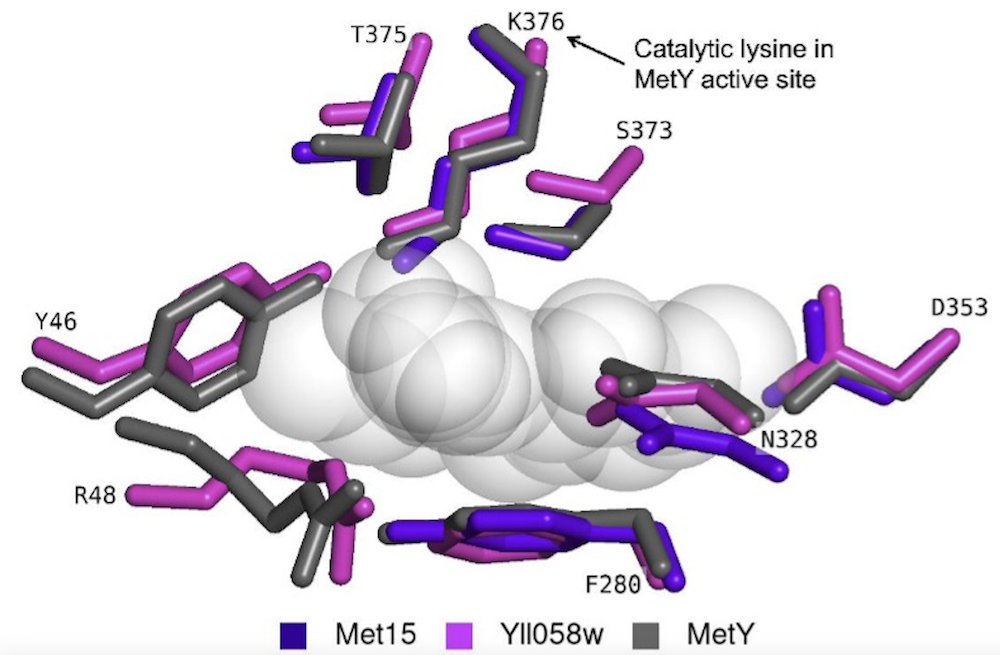 On the illusion of auxotrophy: met15Δ yeast cells can grow on inorganic sulfur thanks to the previously uncharacterized homocysteine synthase Yll058w
On the illusion of auxotrophy: met15Δ yeast cells can grow on inorganic sulfur thanks to the previously uncharacterized homocysteine synthase Yll058w
Van Oss SB, Parikh SB, Castilho Coelho N, Wacholder A, Belashov I, Zdancewicz S, Michaca M, Xu J, Kang YP, Ward NP, Yoon SJ, McCourt KM, McKee J, Ideker T, VanDemark AP, DeNicola GM, Carvunis AR.
Journal of Biological Chemistry (2022)
Link to spotlight in ASBMB Today.
Origins, evolution, and physiological implications of de novo genes in yeast
Parikh SB, Houghton C, Van Oss SB, Wacholder A, Carvunis AR
Yeast (2022)
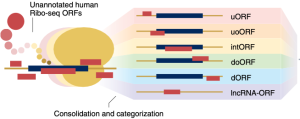 Standardized annotation of translated open reading frames
Standardized annotation of translated open reading frames
Jonathan M. Mudge, Jorge Ruiz-Orera, John R. Prensner, Marie A. Brunet, Ferriol Calvet, Irwin Jungreis, Jose Manuel Gonzalez, Michele Magrane, Thomas F. Martinez, Jana Felicitas Schulz, Yucheng T. Yang, M. Mar Albà, Julie L. Aspden, Pavel V. Baranov, Ariel A. Bazzini, Elspeth Bruford, Maria Jesus Martin, Lorenzo Calviello, Anne-Ruxandra Carvunis, Jin Chen, Juan Pablo Couso, Eric W. Deutsch, Paul Flicek, Adam Frankish, Mark Gerstein, Norbert Hubner, Nicholas T. Ingolia, Manolis Kellis, Gerben Menschaert, Robert L. Moritz, Uwe Ohler, Xavier Roucou, Alan Saghatelian, Jonathan S. Weissman, & Sebastiaan van Heesch
Nature Biotechnology (2022)
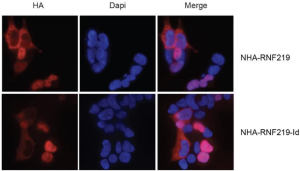 RNF219 regulates CCR4-NOT function in mRNA translation and deadenylation
RNF219 regulates CCR4-NOT function in mRNA translation and deadenylation
Guénolé A, Velilla F, Chartier A, Rich A, Carvunis AR, Sardet C, Simonelig M, Sobhian B.
Scientific Reports (2022)
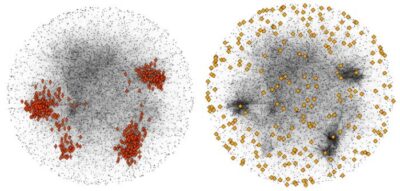 Elastic Network Modeling of Cellular Networks Unveils Sensor and Effector Genes that Control Information Flow
Elastic Network Modeling of Cellular Networks Unveils Sensor and Effector Genes that Control Information Flow
Acar O, Zhang S, Bahar I, Carvunis AR.
Plos Computational Biology (2022)
 Evolutionary characterization of the short protein SPAAR
Evolutionary characterization of the short protein SPAAR
Lee J, Wacholder A, Carvunis AR.
Genes (2021)
 Li Detector: a framework for sensitive colony-based screens regardless of the distribution of fitness effects
Li Detector: a framework for sensitive colony-based screens regardless of the distribution of fitness effects
Parikh SB, Castilho Coelho N, Carvunis AR.
G3 (2021)
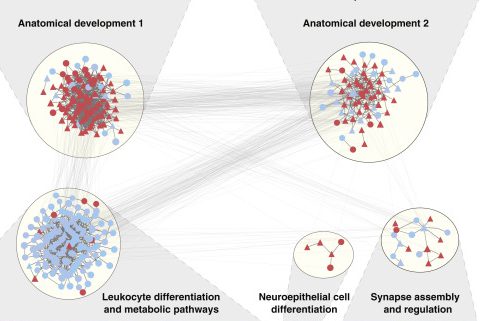 Quantitative Translation of Dog-to-Human Aging by Conserved Remodeling of the DNA Methylome.
Quantitative Translation of Dog-to-Human Aging by Conserved Remodeling of the DNA Methylome.
Wang T, Ma J, Hogan AN, Fong S, Licon K, Tsui B, Kreisberg JF, Adams PD, Carvunis AR, Bannasch DL, Ostrander EA, Ideker T.
Cell Systems (2020).
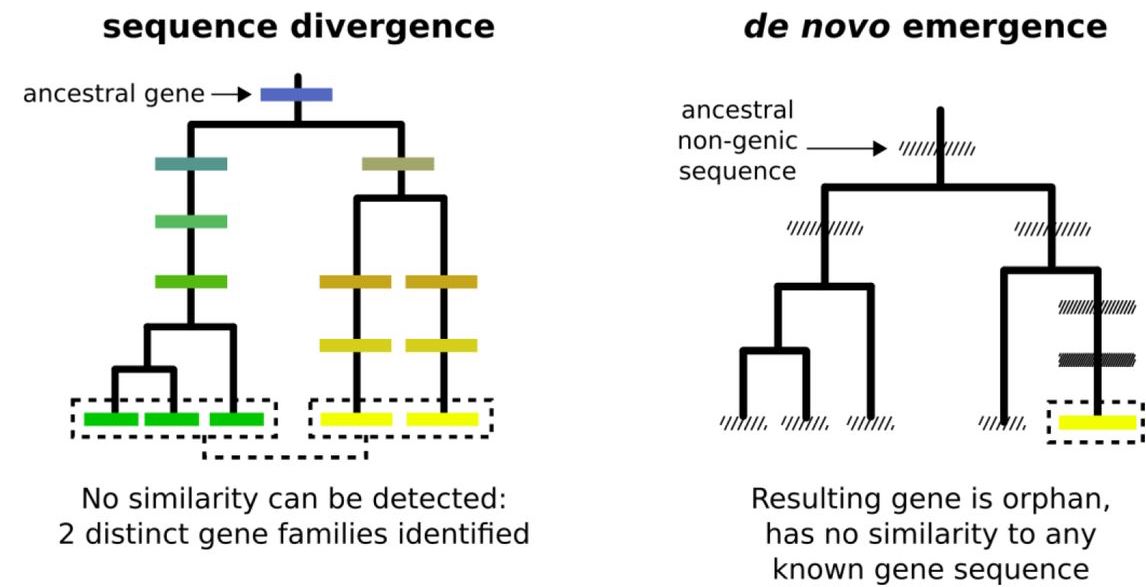 Synteny-based analyses indicate that sequence divergence is not the main source of orphan genes.
Synteny-based analyses indicate that sequence divergence is not the main source of orphan genes.
Vakirlis N, Carvunis AR*, McLysaght A*. *=corresponding authors
eLIFE (2020).
Link to spotlight in eLIFE.
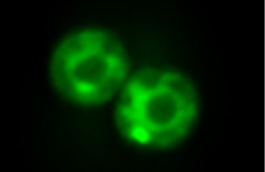 De novo emergence of adaptive membrane proteins from thymine-rich genomic sequences.
De novo emergence of adaptive membrane proteins from thymine-rich genomic sequences.
Vakirlis N, Acar O, Hsu B, Castilho Coelho N, Van Oss SB, Wacholder A, Medetgul-Ernar K, Bowman II RW, Hines CP, Iannotta J, Parikh SB, McLysaght A, Camacho CJ, O’Donnell AF*, Ideker T*, Carvunis AR*. *=corresponding authors
Nature Communications (2020).
Recommended on F1000.
 The Recalcitrance and Resilience of Scientific Function.
The Recalcitrance and Resilience of Scientific Function.
Keeling DM, Garza P, Nartey CM, Carvunis AR.
Poroi (2020).
 The meanings of ‘function’ in biology and the problematic case of de novo gene emergence.
The meanings of ‘function’ in biology and the problematic case of de novo gene emergence.
Keeling DM, Garza P, Nartey CM*, Carvunis AR*. *=corresponding authors
eLife (2019).
Recommended on F1000.
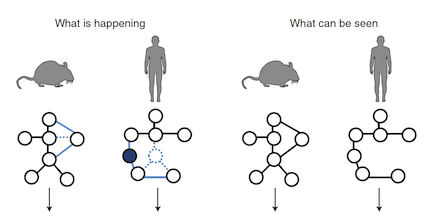 Of mice, men and immunity: a case for evolutionary systems biology.
Of mice, men and immunity: a case for evolutionary systems biology.
Ernst P, Carvunis AR.
Nature Immunology (2018).
 No evidence for phylostratigraphic bias impacting inferences on patterns of gene emergence and evolution.
No evidence for phylostratigraphic bias impacting inferences on patterns of gene emergence and evolution.
Domazet-Lošo T*, Carvunis AR*,#, Albà MM, Šestak MS, Bakarić R, Neme R, Tautz D#. *=co-first authors. #=co-corresponding authors.
MBE (2017).
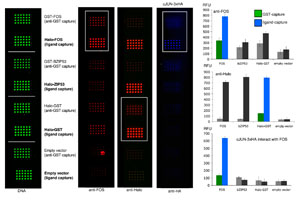
Mapping transcription factor interactome networks using HaloTag protein arrays.
Yazakia J, Galli M, Kim AY, Nito K, Aleman F, Chang KN, Carvunis AR, Quan R, Nguyen H, Song L, Alvarez JM, Huang SC, Chen H, Ramachandran N, Altmann S, Gutiérrez RA, Hill DE, Schroederd JI, Chory J, LaBaer J, Vidal M, Braun P, Ecker JR
PNAS (2016).
 Evidence for a common evolutionary rate in metazoan transcriptional networks.
Evidence for a common evolutionary rate in metazoan transcriptional networks.
Carvunis AR*, Wang T*, Skola D*, Yu A, Chen J, Kreisberg J, Ideker T. *=co-first authors.
Elife (2015).
 A proteome-scale map of the human interactome network.
A proteome-scale map of the human interactome network.
Rolland T*, Tasan M*, Charloteaux B*, Pevzner SJ*, Zhong Q*, Sahni N*, Yi S*, Lemmens I, Fontanillo C, Mosca R, Kamburov A, Ghiassian S, Yang X, Ghamsari L, Balcha D, Begg BE, Braun P, Brehme M, Broly MP, Carvunis AR, Convery-Zupan D, Corominas R, Hardy MF, Coulombe-Huntington J, Dann E, Dreze M, Dricot A, Fan C, Franzosa E, Gebreab F, Gutierrez BJ, Jin M, Kang S, Kiros R, Lin GN, Luck K, MacWilliams A, Menche J, Murray RR, Palagi A, Poulin MM, Rambout X, Rasla J, Reichert P, Romero V, Ruyssinck E, Sahalie J, Scholz A, Shah AA, Sharma A, Shen Y, Spirohn K, Tam S, Tejeda AO, Trigg SA, Twizere JC, Vega K, Walsh J, Cusick ME, Xia Y, Barabási AL, Iakoucheva LM, Aloy P, De Las Rivas J, Tavernier J, Calderwood MA, Hill DE, Hao T, Roth FP, Vidal M. *=co-first authors.
Cell (2014). Recommended on F1000.
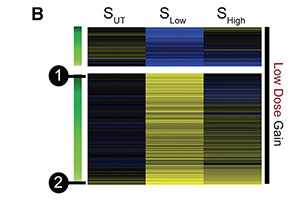 A UV-induced genetic network links the RSC complex to nucleotide excision repair and shows dose-dependent rewiring.
A UV-induced genetic network links the RSC complex to nucleotide excision repair and shows dose-dependent rewiring.
Srivas R*, Costelloe T*, Carvunis AR, Sarkar S, Malta E, Sun SM, Pool M, Licon K, van Welsem T, van Leeuwen F, McHugh PJ, van Attikum H, Ideker T. *=co-first authors.
Cell Reports (2013).
 Integrative approaches for finding modular structure in biological networks.
Integrative approaches for finding modular structure in biological networks.
Mitra K*, Carvunis AR*, Ramesh SK, Ideker T. *=co-first authors.
Nature Reviews Genetics (2013).
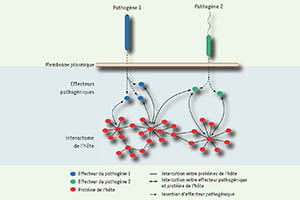 Les facteurs de virulence ciblent des protéines clés de l’interactome de l’hôte (Virulence effectors target key proteins of interactome networks of host plant cells).
Les facteurs de virulence ciblent des protéines clés de l’interactome de l’hôte (Virulence effectors target key proteins of interactome networks of host plant cells).
Carvunis AR, Dreze M
Médecine/Sciences (2012).
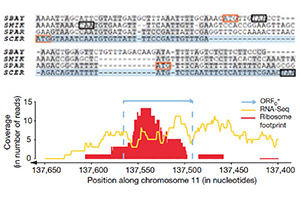 Proto-genes and de novo gene birth.
Proto-genes and de novo gene birth.
Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, Brar GA, Weissman JS, Regev A, Thierry-Mieg N, Cusick ME, Vidal M
Nature (2012).
Recommended on F1000.
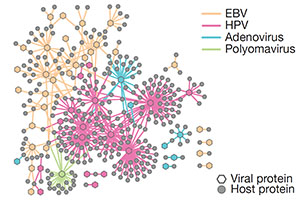 Interpreting cancer genomes using systematic host perturbations by tumour virus proteins.
Interpreting cancer genomes using systematic host perturbations by tumour virus proteins.
Rozenblatt-Rosen O*, Deo RC*, Megha Padi*, Adelmant G*, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares T, Pevzner S, Abderazzaq F, Byrdsong D, Chen A, Cheng J, Correll M, Duarte M, Fan C, Ficarro SB, Franchi R, Garg B, Gulbahce N, Holthaus A, James R, Korkhin A, Litovchick L, Mar JC, Pak TR, Rabello S, Rubio R, Shen Y, Singh S, Spangle JM, Tasan M, Wanamaker S, Webber JT, Carvunis AR, Roecklein-Canfield J, Johannsen E, Barabási AL, Beroukhim R, Kieff E, Cusick ME, Hao T, Hill DE, Münger K, Marto JA, Quackenbush J, Roth FR, DeCaprio JA, Vidal M. *=co-first authors.
Nature (2012).
 Genome-Wide Identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model.
Genome-Wide Identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model.
Feinbaum RL, Urbach JM, Liberati NT, Djonovic S, Adonizio A, Carvunis AR, Ausubel FM
PLoS Pathogens (2012).
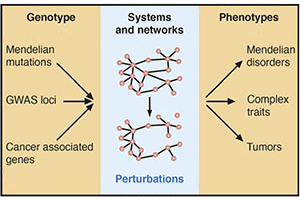 Interactome networks.
Interactome networks.
Carvunis AR, Roth FP, Calderwood MA, Cusick ME, Superti-Furga G, Vidal M
Handbook of Systems Biology (2012).
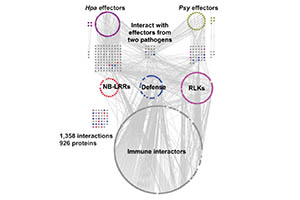 Independently Evolved Virulence Effectors Converge onto Cellular Hubs in a Plant Immune System Network.
Independently Evolved Virulence Effectors Converge onto Cellular Hubs in a Plant Immune System Network.
Mukhtar MS*, Carvunis AR*, Dreze M*, Epple P*, Steinbrener J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, Pevzner SJ, Donovan SE, Ghamsari L, Balaji S, Romero V, Poulin MM, Gebreab F, Gutierrez BJ, Tam S, Harbort C, McDonald N, Gai L, Chen H, EU Effectoromics Consortium, Roth FP, Ecker JR, Vidal M, Beynon J, Braun P, Dangl J. *=co-first authors.
Science (2011).
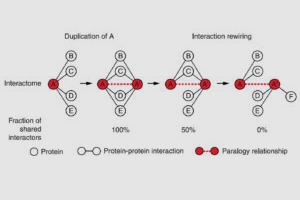 Evidence for Network Evolution in an Arabidopsis Interactome Map.
Evidence for Network Evolution in an Arabidopsis Interactome Map.
Arabidopsis Interactome Mapping Consortium (Dreze M*, Carvunis AR*, Charloteaux B*, Galli M*, Pevzner S*, Tasan M. *=co-first authors.)
Science (2011)
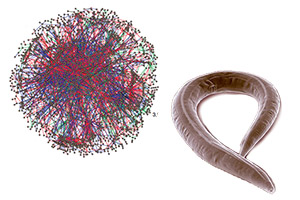 Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network.
Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network.
Simonis N*, Rual JF*, Carvunis AR*, Tasan M*, Lemmens I*, Hirozane-Kishikawa T, Hao T, Sahalie JM, Venkatesan K, Gebreab F, Cevik S, Klitgord N, Fan C, Braun P, Li N, Ayivi- Guedehoussou N, Dann E, Bertin N, Szeto D, Dricot A, Yildirim MA, Lin C, de Smet AS, Kao HL, Simon C, Smolyar A, Ahn JS, Tewari M, Boxem M, Milstein S, Yu H, Dreze M, Vandenhaute J, Gunsalus KC, Cusick ME, Hill DE, Tavernier J, Roth FP, Vidal M. *=co-first authors.
Nature Methods (2009).
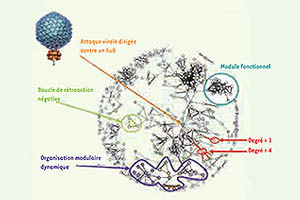 Biologie systémique: des concepts d’hier aux découvertes de demain (Systems biology: from yesterday’s concepts to tomorrow’s discoveries).
Biologie systémique: des concepts d’hier aux découvertes de demain (Systems biology: from yesterday’s concepts to tomorrow’s discoveries).
Carvunis AR, Gómez E, Thierry-Mieg N, Trilling L, Vidal M
Médecine/Sciences (2009).
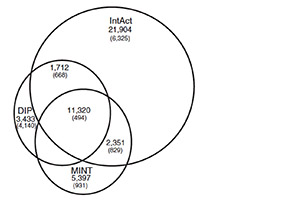 Literature-curated protein interaction datasets.
Literature-curated protein interaction datasets.
Cusick ME*, Yu H*, Smolyar A, Venkatesan K, Carvunis AR, Simonis N, Rual JF, Borick H, Braun P, Dreze M, Vandenhaute J, Galli M, Yazaki J, Hill DE, Ecker JR, Roth FP, Vidal M. *=co-first authors.
Nature Methods (2009).
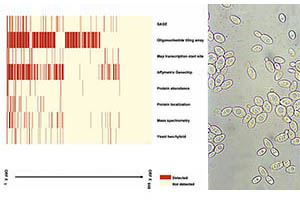 Revisiting the Saccharomyces cerevisiae predicted ORFeome.
Revisiting the Saccharomyces cerevisiae predicted ORFeome.
Li QR*, Carvunis AR*, Yu H*, Han JD*, Zhong Q, Simonis N, Tam S, Hao T, Klitgord NJ, Dupuy D, Mou D, Wapinski I, Regev A, Hill DE, Cusick ME, Vidal M. *=co-first authors.
Genome Research (2008).
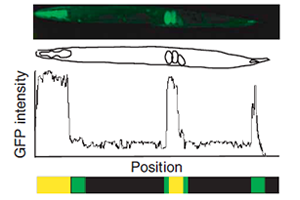 Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans..
Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans..
Dupuy D*, Bertin N*, Hidalgo CA*, Venkatesan K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, Carvunis AR, Pulak R, Shingles J, Reece-Hoyes J, Hunt-Newbury R, Viveiros R, Mohler WA, Tasan M, Roth FP, Le Peuch C, Hope IA, Johnsen R, Moerman DG, Barabási AL, Baillie D, Vidal M. *=co-first authors.
Nature Biotechnology (2007).
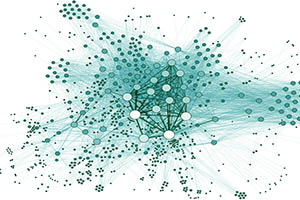 Dynamics of three-state excitable units on Poisson versus power-law random networks.
Dynamics of three-state excitable units on Poisson versus power-law random networks.
Carvunis AR, Latapy M, Lesne A, Magnien C, Pezard L
Physica A (2006).

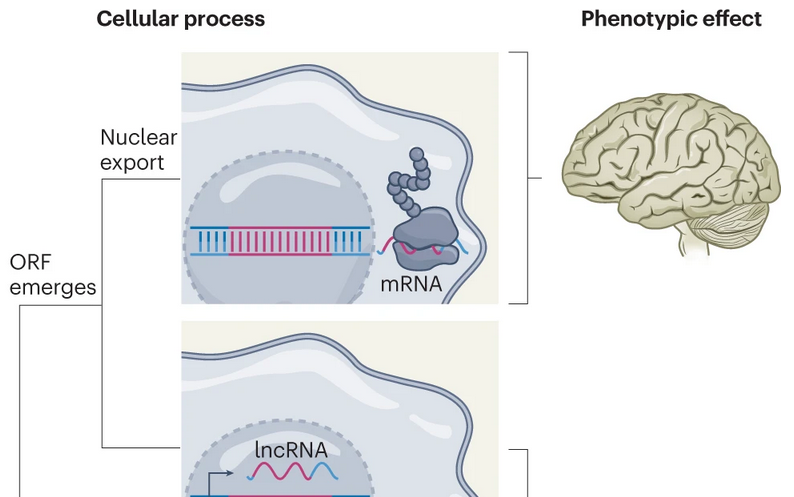 De novo gene increases brain size
De novo gene increases brain size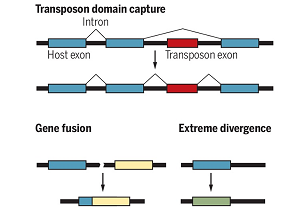 New genes from borrowed parts
New genes from borrowed parts